Diagnostics and association of Alzheimer’s disease markers with metabolomic age.
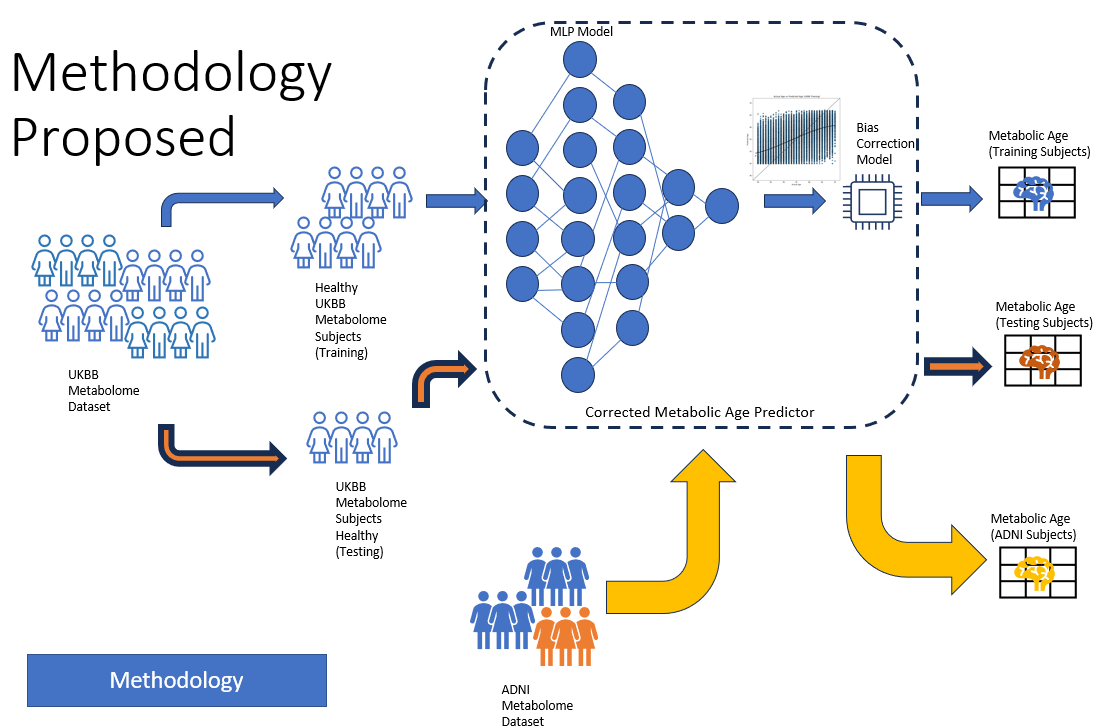
My academic research is based on understanding Alzheimer’s disease pathology, including building the best diagnostic tools for AD categorization and also on how these tools and methodologies can work on other chronic conditions such as cancer, diabetes and so.
Alzheimer’s disease (AD) is one of the most common form of brain dementia, along with which substantial failure of organs and mental issues arise. The heritability of AD is very high, estimated to be between 60% and 80% with much requirement to be done in investigating the genetic aspect of the pathology. With the burst of sequencing and advancement of multiple branches of omics, the amount and variety of data that can be collected for a single study also collectively increased.
Starting with Genome-Wide Association Studies (GWAS) that are able to calculate the most significantly associated variant to the phenotype, expression quantitative trait loci (eQTL), which provides a region of genome that have variations and correlate with gene expression, and RNA-sequencing data which provides the exact value of expression of the gene at a given point of time from their RNA composition, belong just in the genomic division of dataset. Having such closely related datasets, I can take a step back to further understand how the variants affect or cause the pathology of AD. Further, the epigenomic functional analysis of these variants can allow us to look at how the variants are being associated to the gene and the disease. I also incorporate the metabolomics and imaging studies to calculate the how a multi-omics approach can help identifying variants and genes that explain pathology of AD. The collective aim of my research is to identify the functional variants and metabolite effect to construct a continuous multi-omics-based pathology for AD.